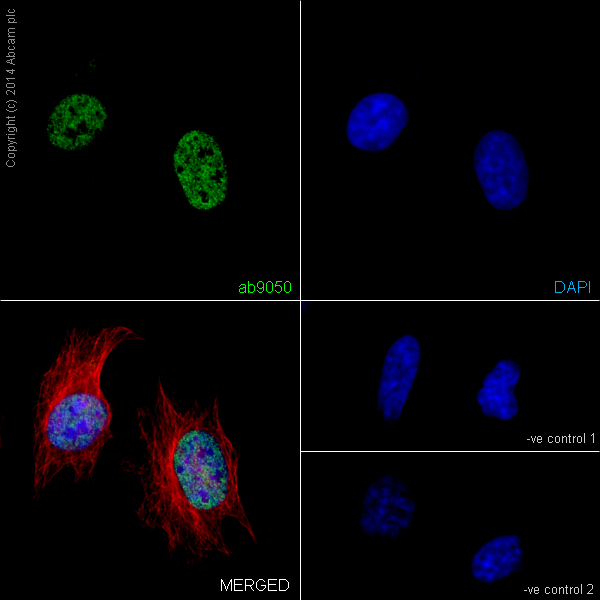
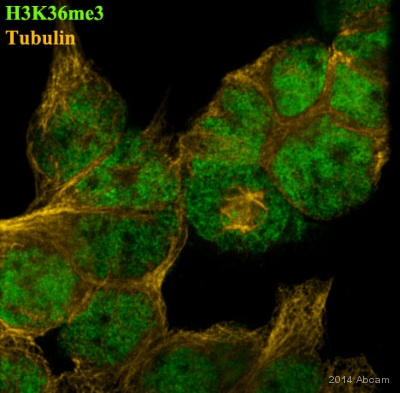
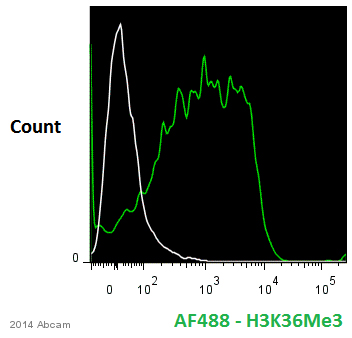
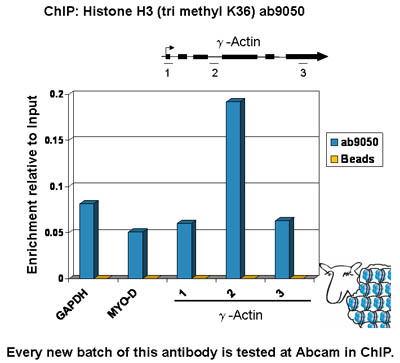
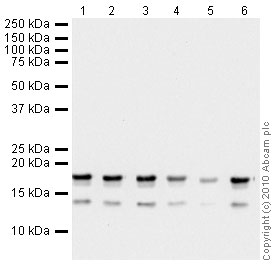
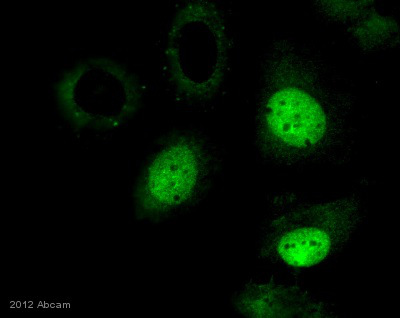
Anti-Histone H3 (tri methyl K36) antibody - ChIP Grade
| Name | Anti-Histone H3 (tri methyl K36) antibody - ChIP Grade |
|---|---|
| Supplier | Abcam |
| Catalog | ab9050 |
| Prices | $404.00 |
| Sizes | 100 µg |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
| Applications | ICC/IF ICC/IF ChIPseq WB ChIP ChIP ChIP IHC-P |
| Species Reactivities | Mouse, Rat, Bovine, Human, Yeast, Xenopus, Arabidopsis thaliana, C. elegans, Drosophila, S. pombe, Zebrafish, Rice, Plant |
| Antigen | Synthetic peptide conjugated to KLH derived from within residues 1 - 100 of Human Histone H3, tri methylated at K36 |
| Blocking Peptide | Human Histone H3 (tri methyl K36) peptide |
| Description | Rabbit Polyclonal |
| Gene | HIST3H3 |
| Conjugate | Unconjugated |
| Supplier Page | Shop |
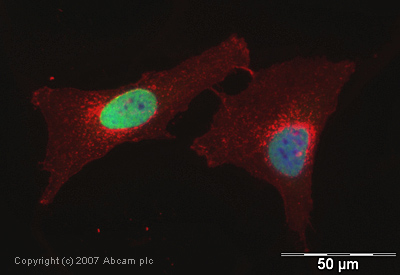
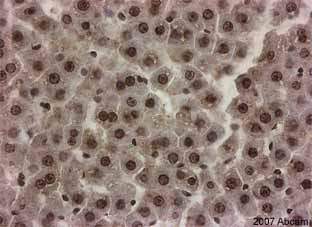
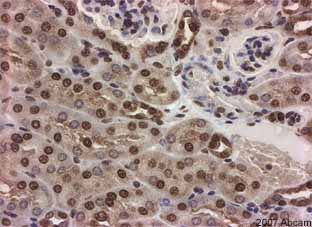
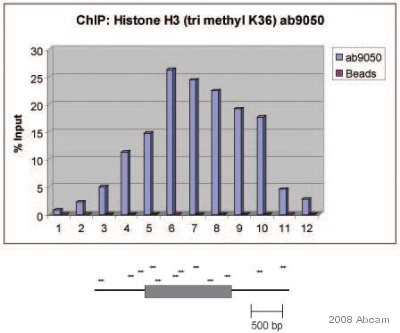
Product images
Product References
The hematopoietic regulator TAL1 is required for chromatin looping between the - The hematopoietic regulator TAL1 is required for chromatin looping between the
Yun WJ, Kim YW, Kang Y, Lee J, Dean A, Kim A. Nucleic Acids Res. 2014 Apr;42(7):4283-93.
Regulation of MYC expression and differential JQ1 sensitivity in cancer cells. - Regulation of MYC expression and differential JQ1 sensitivity in cancer cells.
Fowler T, Ghatak P, Price DH, Conaway R, Conaway J, Chiang CM, Bradner JE, Shilatifard A, Roy AL. PLoS One. 2014 Jan 23;9(1):e87003.
Variation in chromatin accessibility in human kidney cancer links H3K36 - Variation in chromatin accessibility in human kidney cancer links H3K36
Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS, Prins JF, Lieb JD, Rathmell WK, Davis IJ. Genome Res. 2014 Feb;24(2):241-50.
Canonical nucleosome organization at promoters forms during genome activation. - Canonical nucleosome organization at promoters forms during genome activation.
Zhang Y, Vastenhouw NL, Feng J, Fu K, Wang C, Ge Y, Pauli A, van Hummelen P, Schier AF, Liu XS. Genome Res. 2014 Feb;24(2):260-6.
Systematic mapping of occluded genes by cell fusion reveals prevalence and - Systematic mapping of occluded genes by cell fusion reveals prevalence and
Looney TJ, Zhang L, Chen CH, Lee JH, Chari S, Mao FF, Pelizzola M, Zhang L, Lister R, Baker SW, Fernandes CJ, Gaetz J, Foshay KM, Clift KL, Zhang Z, Li WQ, Vallender EJ, Wagner U, Qin JY, Michelini KJ, Bugarija B, Park D, Aryee E, Stricker T, Zhou J, White KP, Ren B, Schroth GP, Ecker JR, Xiang AP, Lahn BT. Genome Res. 2014 Feb;24(2):267-80.
D-2-hydroxyglutarate produced by mutant IDH2 causes cardiomyopathy and - D-2-hydroxyglutarate produced by mutant IDH2 causes cardiomyopathy and
Akbay EA, Moslehi J, Christensen CL, Saha S, Tchaicha JH, Ramkissoon SH, Stewart KM, Carretero J, Kikuchi E, Zhang H, Cohoon TJ, Murray S, Liu W, Uno K, Fisch S, Jones K, Gurumurthy S, Gliser C, Choe S, Keenan M, Son J, Stanley I, Losman JA, Padera R, Bronson RT, Asara JM, Abdel-Wahab O, Amrein PC, Fathi AT, Danial NN, Kimmelman AC, Kung AL, Ligon KL, Yen KE, Kaelin WG Jr, Bardeesy N, Wong KK. Genes Dev. 2014 Mar 1;28(5):479-90.
The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites - The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites
Pedersen MT, Agger K, Laugesen A, Johansen JV, Cloos PA, Christensen J, Helin K. Mol Cell Biol. 2014 Mar;34(6):1031-45.
Sudemycin E influences alternative splicing and changes chromatin modifications. - Sudemycin E influences alternative splicing and changes chromatin modifications.
Convertini P, Shen M, Potter PM, Palacios G, Lagisetti C, de la Grange P, Horbinski C, Fondufe-Mittendorf YN, Webb TR, Stamm S. Nucleic Acids Res. 2014 Apr;42(8):4947-61.
Genome-wide analysis of the chromatin composition of histone H2A and H3 variants - Genome-wide analysis of the chromatin composition of histone H2A and H3 variants
Yukawa M, Akiyama T, Franke V, Mise N, Isagawa T, Suzuki Y, Suzuki MG, Vlahovicek K, Abe K, Aburatani H, Aoki F. PLoS One. 2014 Mar 21;9(3):e92689.
Effects of RNAi-mediated knockdown of histone methyltransferases on the - Effects of RNAi-mediated knockdown of histone methyltransferases on the
Suzuki MG, Ito H, Aoki F. Int J Mol Sci. 2014 Apr 22;15(4):6772-96.